Searchlight classification of movie segments
Contents
Searchlight classification of movie segments#
In this analysis, we classify which segment (5 TRs) of the movie the participant was watching for each searchlight (20 mm radius).
Preparations#
Import Python packages#
import os
import numpy as np
from scipy.stats import ttest_rel, rankdata, pearsonr, spearmanr
import neuroboros as nb
from joblib import Parallel, delayed, cpu_count, parallel_backend
import matplotlib.pyplot as plt
import matplotlib as mpl
import seaborn as sns
Other settings#
dsets = ['raiders', 'budapest']
ico = 32
spaces = [f'{a}-ico{b}' for a in ['fsavg', 'fslr', 'onavg'] for b in [ico]]
train_dur, test_dur = '1st-half', '2nd-half'
radius = 20
align_radius = 20
size = 5
colors = np.array(sns.color_palette('tab10'))
np.set_printoptions(4, linewidth=200)
if os.path.exists('Arial.ttf'):
from matplotlib import font_manager
font_manager.fontManager.addfont('Arial.ttf')
mpl.rcParams['font.family'] = 'Arial'
Load results#
# cache file to avoid excessive I/O
summary_fn = f'sl_clf/summary_ico{ico}_size{size}.pkl'
if not os.path.exists(summary_fn):
results = {}
for dset_name in dsets:
dset = nb.dataset(dset_name)
sids = dset.subjects
for align in ['surf', 'procr', 'ridge']:
for space in spaces:
folder = f'sl_clf/{dset_name}/{space}_{align}_{align_radius}mm_'\
f'{train_dur}_{test_dur}_{size}TRs'
aa = []
for sid in sids:
a = [np.load(f'{folder}/{sid}_{lr}h_{radius}mm.npy')
for lr in 'lr']
a = np.concatenate(a)
aa.append(a)
aa = np.array(aa)
results[dset_name, space, align] = aa
nb.save(summary_fn, results)
results = nb.load(summary_fn)
Accuracy of movie time point classification#
Two datasets (top: Raiders, bottom: Budapest)
Three alignment methods (surface alignment, Procrustes hyperalignment, warp hyperalignment)
Bars denote average accuracy across participants and searchlights
Grey lines denote individual participants (average accuracy across searchlights)
fig, axs = plt.subplots(2, 3, figsize=[_/2.54 for _ in (12, 8)], dpi=200)
pct1, pct2, pp = [], [], []
for ii, dset_name in enumerate(['raiders', 'budapest']):
for jj, align in enumerate(['surf', 'procr', 'ridge']):
ax = axs[ii, jj]
ys = []
for i, space in enumerate(spaces):
a = results[dset_name, space, align].mean(axis=1)
m = a.mean(axis=0)
c = colors[i]
ax.bar(i, m, facecolor=c, edgecolor=c*0.5, ecolor=c*0.5)
ys.append(a)
ax.set_xticks(np.arange(3), labels=spaces)
ys = np.array(ys)
for y in ys.T:
ax.plot(np.arange(3), y, 'k-', alpha=0.3, markersize=1)
t, p = ttest_rel(ys[2], ys[0])
pp.append(p)
assert p < 0.005
t, p = ttest_rel(ys[2], ys[1])
pp.append(p)
assert p < 0.005
pct1.append(np.mean(ys[0] < ys[2]))
pct2.append(np.mean(ys[1] < ys[2]))
max_ = int(np.ceil(ys.max() * 20))
yy = np.arange(max_ + 1) * 0.05
ax.set_yticks(yy, labels=[f'{_*100:.0f}%' for _ in yy])
ax.set_xlim([-0.6, 2.6])
ax.tick_params(axis='both', pad=0, length=2, labelsize=5)
ax.set_ylabel('Accuracy', size=6, labelpad=1)
ax.set_xlabel('Surface space', size=6, labelpad=1)
title = {'procr': 'Procrustes hyperalignment',
'ridge': 'Warp hyperalignment',
'surf': 'Surface alignment'}[align]
ax.set_title(f'{title}', size=6, pad=2)
ax.annotate(dset_name.capitalize(), (0.005, 0.99 - ii * 0.5),
xycoords='figure fraction', size=6, va='top')
print(np.mean(pct1), np.mean(pct2), np.max(pp))
fig.subplots_adjust(
left=0.08, right=0.99, top=0.94, bottom=0.06, wspace=0.3, hspace=0.4)
plt.savefig('sl_clf_bar.png', dpi=300, transparent=True)
plt.show()
1.0 1.0 4.803615811053366e-07

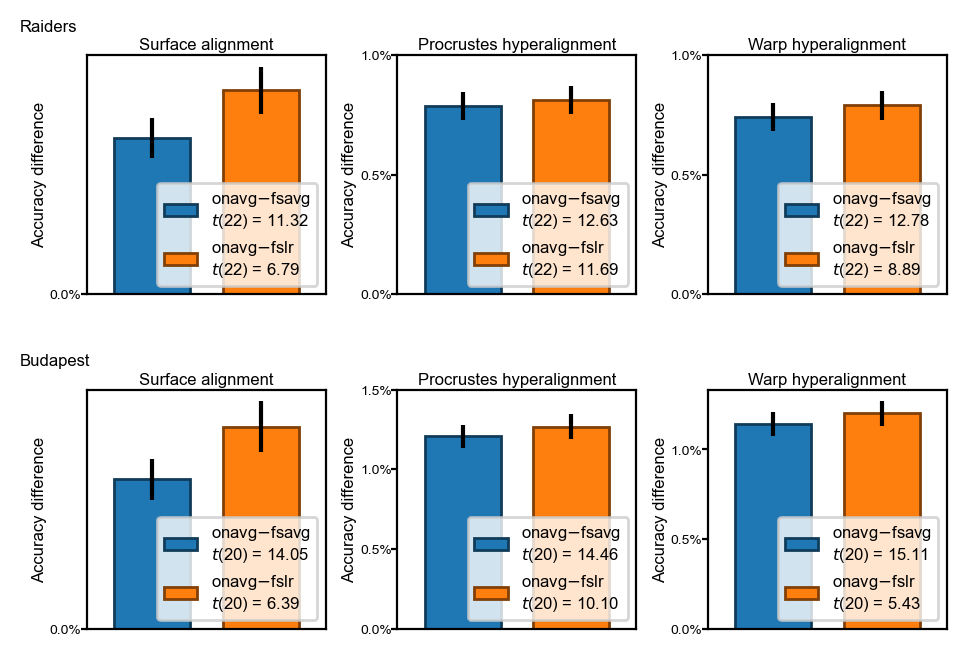
Difference of classification accuracy between template spaces#
The difference accuracy between
onavgand fsavg (blue bars), and betweenonavgand fslr (orange bars)The differences are statistically significant (p < 1e-4) across different datasets and alignment methods
fig, axs = plt.subplots(
2, 3, figsize=[_/2.54 for _ in (12, 8)], dpi=200)
pct1, pct2, pp = [], [], []
for ii, dset_name in enumerate(dsets):
for jj, align in enumerate(['surf', 'procr', 'ridge']):
ax = axs[ii, jj]
max_ = []
onavg = results[dset_name, spaces[-1], align].mean(axis=1)
for kk, space in enumerate(spaces[:2]):
y = onavg - results[dset_name, space, align].mean(axis=1)
m = y.mean(axis=0)
se = y.std(axis=0, ddof=1) / np.sqrt(y.shape[0])
c = colors[kk]
t, p = ttest_rel(onavg, results[dset_name, space, align][:, -1])
dof = len(onavg) - 1
label = f'{spaces[-1].split("-")[0]}$ - ${spaces[kk].split("-")[0]}'\
f'\n$t$({dof}) = {t:.2f}'
ax.bar(kk, m, yerr=se, color=c, ec=c*0.5, width=0.7, label=label)
max_.append(m + se)
assert p < 1e-4
max_ = int(np.ceil(np.max(max_) * 1.1 * 1000))
ax.set_xticks([])
ax.set_xlim(np.array([-0.6, 1.6]))
yy = np.arange(0, max_ + 1, 5)
ax.set_yticks(yy * 0.001, labels=[f'{_*0.1:.1f}%' for _ in yy])
ax.set_ylabel('Accuracy difference', size=6, labelpad=1)
ax.tick_params(axis='both', pad=0, length=2, labelsize=5)
title = {'procr': 'Procrustes hyperalignment',
'ridge': 'Warp hyperalignment',
'surf': 'Surface alignment',
}[align]
ax.set_title(f'{title}', size=6, pad=2)
ax.legend(fontsize=6, fancybox=True, loc='lower right')
axs[ii, 0].annotate(
dset_name.capitalize(), (-0.28, 1.15),
xycoords='axes fraction', size=6, va='top', ha='left')
fig.subplots_adjust(
left=0.08, right=0.99, top=0.93, bottom=0.02, wspace=0.3, hspace=0.4)
plt.savefig('sl_clf_bar_diff.png', dpi=300, transparent=True)
plt.show()
Which brain region has the most improvement#
Upsample the data#
To compare the results based on different template spaces, we upsample the statistics to
onavg-ico128space
upsampled = {}
for ii, dset_name in enumerate(['raiders', 'budapest']):
for jj, align in enumerate(['surf', 'procr', 'ridge']):
for space in spaces:
a = results[dset_name, space, align].mean(axis=0)
center_space = space.split('-')[0] + '-ico32'
xfms = [nb.mapping(lr, center_space, 'onavg-ico128', mask=True)
for lr in 'lr']
nv1 = xfms[0].shape[0]
a = np.concatenate([a[:nv1] @ xfms[0], a[nv1:] @ xfms[1]])
upsampled[dset_name, space, align] = a
upsampled_ivd = {}
for space in spaces:
center_space = space.split('-')[0] + '-ico32'
ivd_lr = []
for lr in 'lr':
coords, faces = nb.geometry('midthickness', lr, center_space)
surf = nb.Surface(coords, faces)
distances = nb.distances(lr, center_space)
mask = nb.mask(lr, space=center_space)
ivd = []
for i, nbrs in enumerate(surf.neighbors):
if mask[i]:
ivd.append(distances[i, nbrs].mean())
ivd_lr.append(np.array(ivd))
values = []
for lr, ivd in zip('lr', ivd_lr):
sls = nb.sls(lr, radius, space=space)
avg_ivd = np.array([ivd[sl].mean() for sl in sls])
xfm = nb.mapping(lr, center_space, 'onavg-ico128', mask=True)
values.append(avg_ivd @ xfm)
upsampled_ivd[space] = np.concatenate(values)
Correlations with improvement in accuracy#
Regions that were previously undersampled are more likely to have more improvement after switching to onavg (
pearsonr(c, d)in the code below).Regions that have higher accuracies based on traditional templates are more likely to have more improvement after switching to onavg (
pearsonr(a, d)in the code below).
for space in spaces[:2]:
for ii, dset_name in enumerate(['raiders', 'budapest']):
for jj, align in enumerate(['surf', 'procr', 'ridge']):
a = upsampled[dset_name, space, align]
b = upsampled[dset_name, spaces[-1], align]
c = upsampled_ivd[space]
d = b - a
print(f'{space:11s}, {dset_name:8s}, {align:5s}, '
f'{pearsonr(c, d)[0]:7.4f}, {pearsonr(a, d)[0]:7.4f}')
fsavg-ico32, raiders , surf , -0.0902, -0.0007
fsavg-ico32, raiders , procr, 0.2385, 0.4536
fsavg-ico32, raiders , ridge, 0.4063, 0.4944
fsavg-ico32, budapest, surf , -0.1182, -0.0443
fsavg-ico32, budapest, procr, 0.2056, 0.3686
fsavg-ico32, budapest, ridge, 0.4501, 0.3976
fslr-ico32 , raiders , surf , -0.1038, 0.0102
fslr-ico32 , raiders , procr, 0.2697, 0.4402
fslr-ico32 , raiders , ridge, 0.4472, 0.5102
fslr-ico32 , budapest, surf , -0.0832, 0.0718
fslr-ico32 , budapest, procr, 0.2601, 0.3916
fslr-ico32 , budapest, ridge, 0.4820, 0.4256
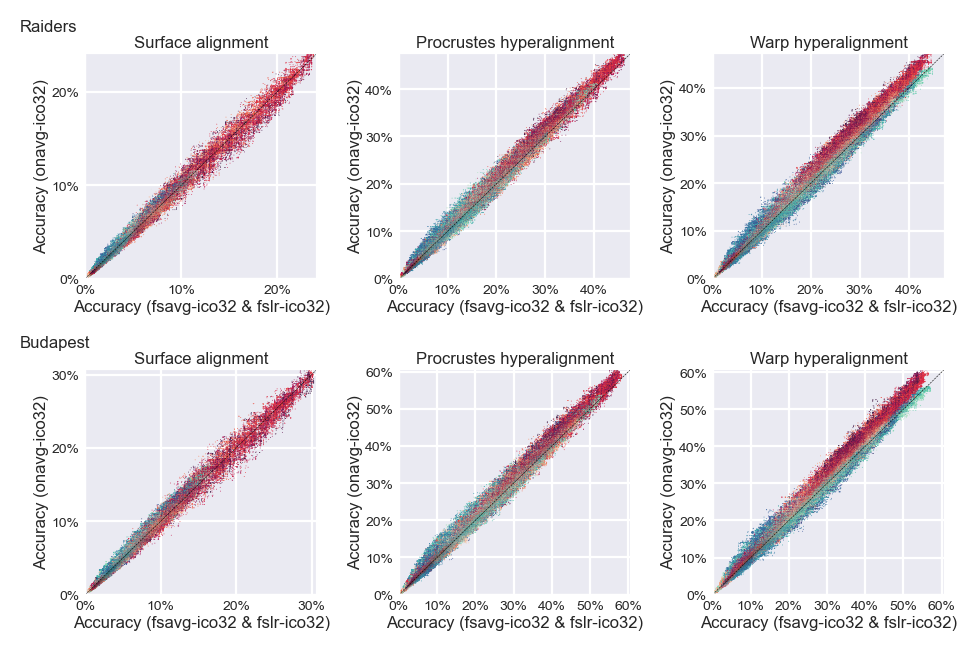
Scatter plots#
nc = 10
colors2 = np.concatenate(
[sns.color_palette("mako", nc)[1:],
[[0.9, 0.9, 0.9]],
sns.color_palette("rocket", nc)[1:][::-1]],
axis=0)
cmap = mpl.colors.LinearSegmentedColormap.from_list('mako_rocket', colors2)
In the figure below, red dots are searchlights that have larger inter-vertex distance based on traditional templates, and blue dots are those that lave smaller inter-vertex distance. Dots above the diagonal are searchlights that improve after switching to onavg, and red dots appear more often above the diagonal.
with sns.axes_style('darkgrid'):
fig, axs = plt.subplots(2, 3, figsize=[_/2.54 for _ in (12, 8)], dpi=200)
for ii, dset_name in enumerate(['raiders', 'budapest']):
for jj, align in enumerate(['surf', 'procr', 'ridge']):
aa, bb, cc = [], [], []
for space in spaces[:2]:
a = upsampled[dset_name, space, align]
b = upsampled[dset_name, spaces[-1], align]
c = upsampled_ivd[space]
aa.append(a)
bb.append(b)
cc.append(c)
aa = np.concatenate(aa)
bb = np.concatenate(bb)
cc = np.concatenate(cc)
cc = cc - np.median(cc)
vmax = np.percentile(cc, [99])
idx = np.argsort(bb - aa)
aa, bb, cc = aa[idx], bb[idx], cc[idx]
ax = axs[ii, jj]
ax.scatter(aa, bb, c=cc, s=0.1, edgecolor='none',
cmap=cmap, vmin=-vmax, vmax=vmax)
lim = [0, max(a.max(), b.max())]
ticks = np.arange(0, np.ceil(lim[1] * 100) + 1, 10) * 0.01
labels = [f'{_*100:.0f}%' for _ in ticks]
ax.set_xticks(ticks, labels=labels)
ax.set_yticks(ticks, labels=labels)
ax.set_xlim(lim)
ax.set_ylim(lim)
ax.plot(lim, lim, 'k--', alpha=1, lw=0.2)
ax.tick_params(axis='both', pad=0, length=2, labelsize=5)
ax.set_ylabel(f'Accuracy ({spaces[-1]})', size=6, labelpad=1)
ax.set_xlabel(f'Accuracy ({" & ".join(spaces[:-1])})',
size=6, labelpad=1)
title = {'procr': 'Procrustes hyperalignment',
'ridge': 'Warp hyperalignment',
'surf': 'Surface alignment',
}[align]
ax.set_title(f'{title}', size=6, pad=2)
axs[ii, 0].annotate(
dset_name.capitalize(), (-0.28, 1.15),
xycoords='axes fraction', size=6, va='top', ha='left')
fig.subplots_adjust(
left=0.08, right=0.99, top=0.92, bottom=0.06, wspace=0.35, hspace=0.4)
plt.savefig('sl_clf_scatter.png', dpi=300)
plt.show()