Different data resolutions
Different data resolutions#
This example is about different input data resolutions.
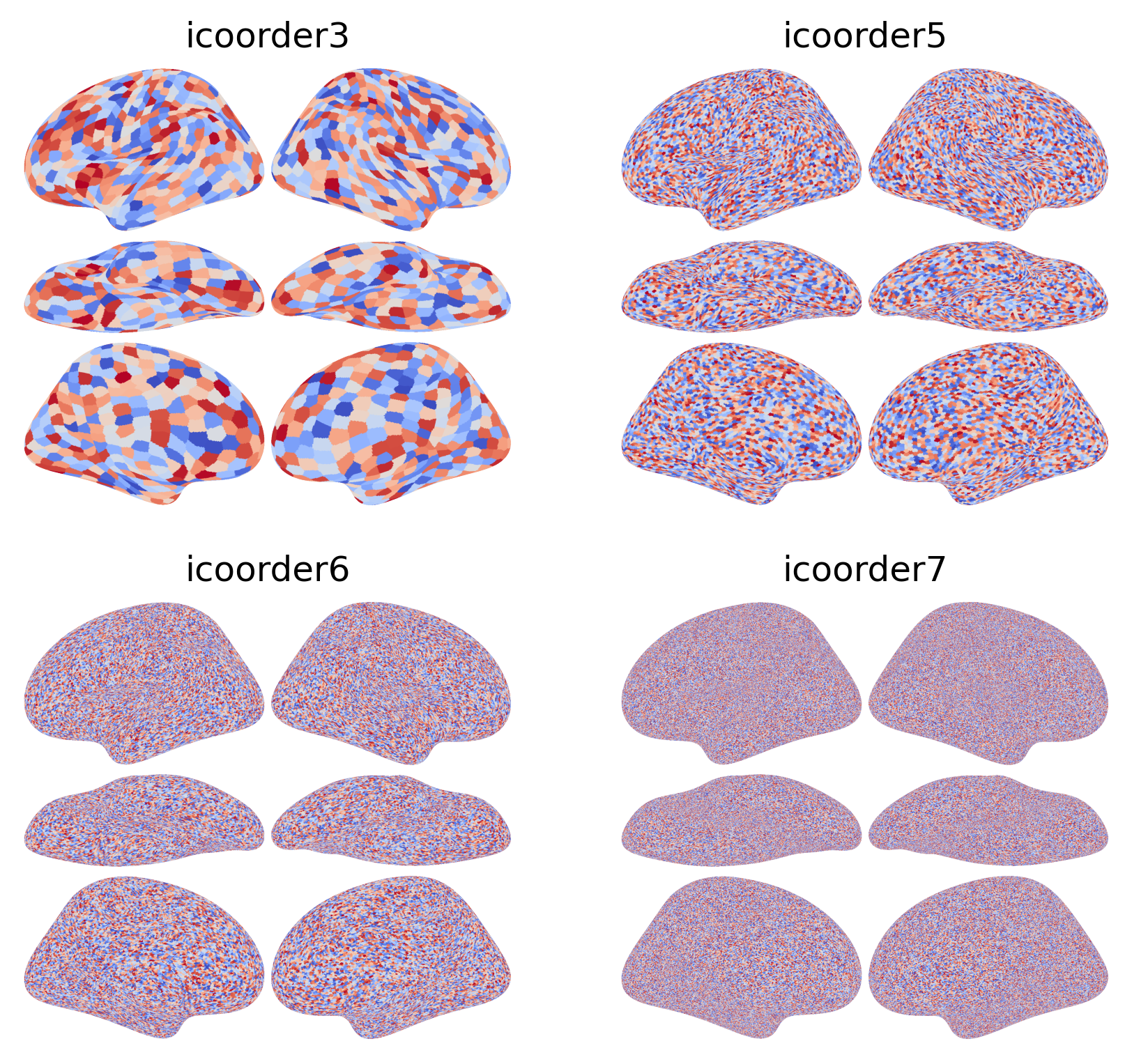
Neuroimaging data on cortical surface has different spatial resolutions:
name |
resolution |
# of vertices per hemishere |
|---|---|---|
icoorder7 |
0.7 mm |
163842 |
icoorder6 |
1.5 mm |
40962 |
icoorder5 |
3 mm |
10242 |
icoorder4 |
6 mm |
2562 |
icoorder3 |
13 mm |
642 |
Typical the data resolution is between icoorder7 (usually anatomical scans) and icoorder5 (usually functional scans). icoorder3 resolution is often used in defining connectivity targets for connectivity hyperalignment and in examples.
The number of cortical vertices per hemisphere can be computed based on icoorder: \( n_v = 4^{icoorder} \times 10 + 2 \), where \(n_v\) is the number of vertices.
The brain_plot function automatically handles various data resolution between icoorder7 and icoorder3. When the input data has a lower resolution than icoorder7, it’s automatically upsampled to the icoorder7 resolution in a nearest-neighbor manner based on the Voronoi diagram. The data is always visualized using the icoorder7 high-resolution surface (i.e., fsaverage).
import numpy as np
from brainplotlib import brain_plot
import matplotlib.pyplot as plt
rng = np.random.default_rng(0)
fig, axs = plt.subplots(2, 2, dpi=300, figsize=([_/300 + 1 for _ in [1728, 1560]]))
icoorders = [3, 5, 6, 7]
for i in range(2):
for j in range(2):
ax = axs[i][j]
icoorder = icoorders[i*2+j]
nv = 4**icoorder * 10 + 2
v = rng.random((nv * 2, ))
img = brain_plot(v, vmax=1, vmin=0, cmap='coolwarm')
ax.imshow(img)
ax.axis('off')
ax.set_title(f'icoorder{icoorder}')
plt.show()

